Abstract
Autoregressive models for molecular graph generation typically operate on flattened sequences of atoms and bonds, discarding the rich multi-scale structure inherent to molecules. We introduce MOSAIC (Multi-scale Organization via Structural Abstraction In Composition), a framework that lifts autoregressive generation from flat token walks to compositional, hierarchy-aware sequences. MOSAIC provides a unified three-stage pipeline: (1) hierarchical coarsening that recursively groups atoms into motif-like clusters using graph-theoretic methods (spectral clustering, hierarchical agglomerative clustering, and motif-aware variants), (2) structured tokenization that serializes the resulting multi-level hierarchy into sequences that explicitly encode parent-child relationships, partition boundaries, and edge connectivity at every level, and (3) autoregressive generation with a standard Transformer decoder that learns to produce these structured sequences. We evaluate MOSAIC on the MOSES and COCONUT molecular benchmarks, comparing four tokenization schemes of increasing hierarchical expressiveness. Our experiments show that hierarchy-aware tokenizations improve chemical validity and structural diversity over flat baselines while enabling control over generated substructures. MOSAIC provides a principled, modular foundation for structure-aware molecular generation.
Introduction
Discovering new molecules — for drugs, materials, or chemical tools — is a slow and expensive process. Recent advances in generative AI offer a way to accelerate this: train a model on known molecules and let it propose entirely new ones. But molecules aren’t just sequences of characters. They have rich internal structure: rings, branches, and recurring building blocks that determine their chemical properties.
Most existing approaches flatten a molecular graph into a linear sequence of atoms and bonds, then generate one token at a time — much like a language model that can only produce text one letter at a time. It technically works, but the model has to rediscover higher-level patterns (like common ring structures) from scratch. MOSAIC is more like generating at the word level: it learns to compose molecules from meaningful building blocks rather than individual atoms.
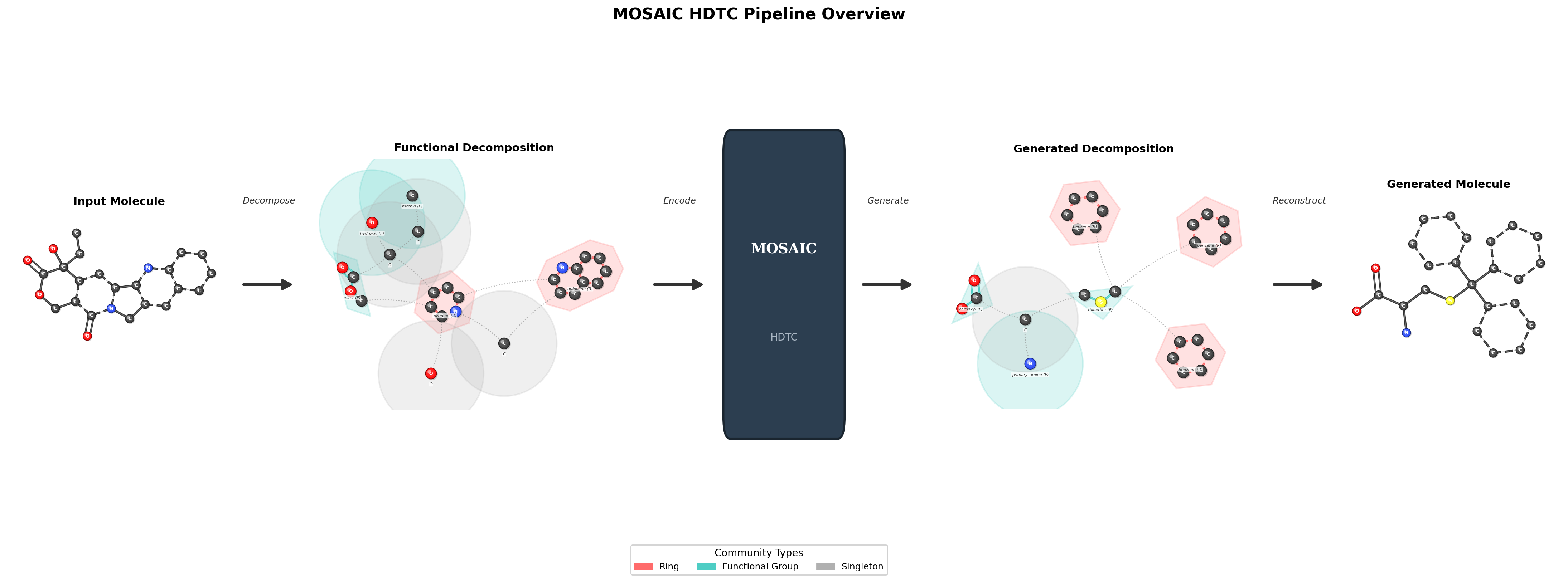
Pipeline Overview
MOSAIC operates through a three-stage pipeline. First, molecular graphs are recursively coarsened into multi-level hierarchies where atoms are grouped into structurally meaningful clusters. Second, these hierarchies are serialized into structured token sequences that preserve parent-child relationships and inter-cluster connectivity. Third, a Transformer decoder is trained to autoregressively generate these structured sequences, enabling the model to compose molecules from coarse structure down to fine-grained atomic detail.
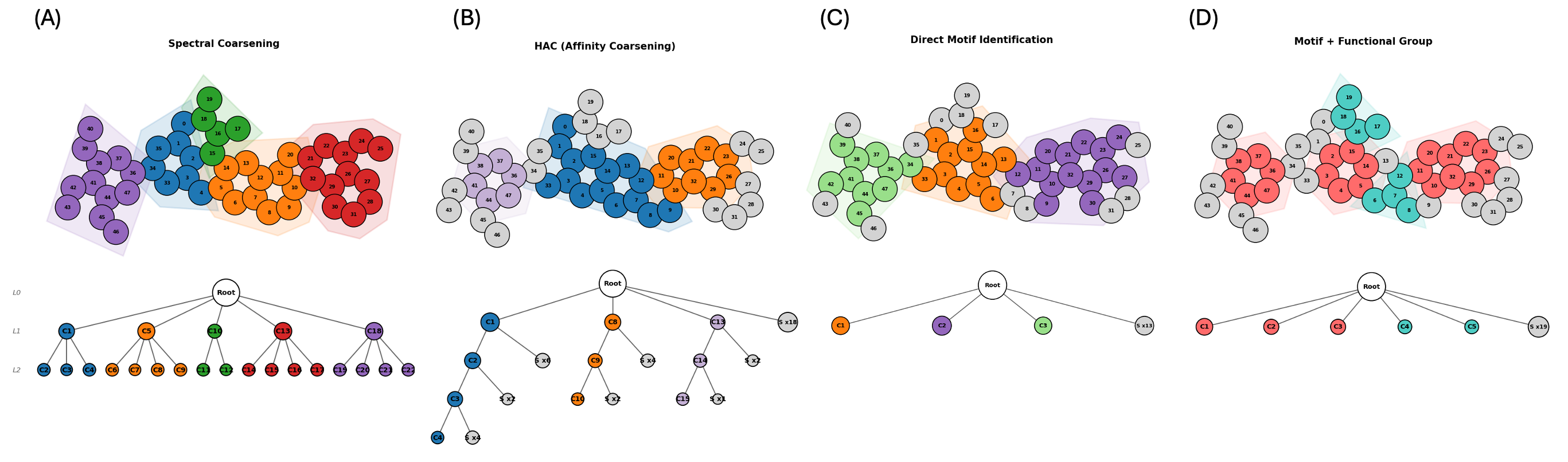
Coarsening Strategies
MOSAIC supports multiple graph coarsening strategies that recursively partition molecular graphs into hierarchical clusters. Each strategy offers different trade-offs between preserving chemical motifs and computational efficiency.
-
Spectral Clustering: Uses the eigenvectors of the graph Laplacian to identify natural clusters in the molecular graph. Spectral methods capture global graph structure and produce balanced partitions.
-
Hierarchical Agglomerative Clustering (HAC): A bottom-up approach that iteratively merges the most similar adjacent nodes based on bond-type-weighted distances. HAC tends to preserve local chemical structure.
-
Motif-aware Coarsening (MC): First identifies known chemical motifs (rings, functional groups) using SMARTS pattern matching, then applies spectral or HAC clustering to the remaining atoms. This ensures that chemically meaningful substructures are preserved as intact units in the hierarchy.
-
Motif-aware + Functional Group (MC+FG): Extends motif-aware coarsening with an expanded library of functional group patterns, providing finer-grained control over which substructures are preserved during coarsening.
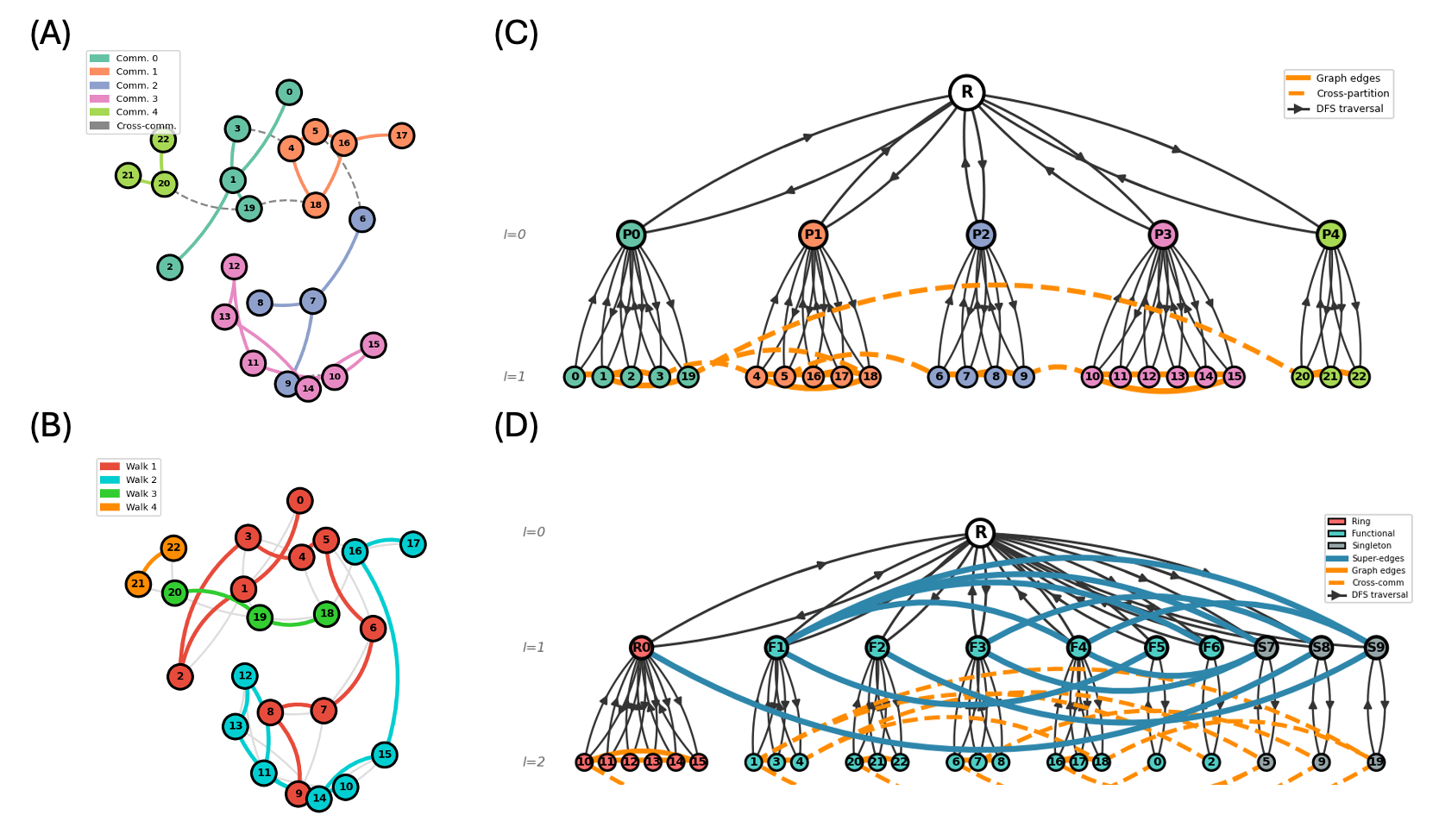
Tokenization Schemes
The coarsened hierarchies are serialized into token sequences using one of four tokenization schemes, each capturing increasing levels of hierarchical information:
-
SENT (Segmented Eulerian Neighborhood Trails): A flat baseline that serializes atom-bond sequences without hierarchy. Serves as the non-hierarchical reference point.
-
H-SENT (Hierarchical SENT): Extends SENT by encoding multi-level partition structure. Tokens include partition boundaries and parent-child relationships, enabling the model to learn coarse-to-fine generation.
-
HDT (Hierarchical Depth Tokenization): Encodes the full tree structure of the hierarchy using depth-first traversal. Each token carries depth information, making the hierarchical nesting explicit.
-
HDTC (Hierarchical Depth Tokenization with Composition): The most expressive scheme, extending HDT with explicit compositional tokens at each internal node. This gives the model complete information about the branching structure of the hierarchy.
Results on COCONUT
We evaluate unconditional generation on the COCONUT dataset of complex natural products (~5K molecules, 30–100 heavy atoms, ≥4 rings). All models use a GPT-2 backbone (12 layers, 768 hidden, 12 heads) trained with identical hyperparameters. We generate 500 molecules per model and compare against the full reference dataset. Bold = best, underline = second best.
| SENT | H-SENT MC | H-SENT SC | H-SENT HAC | HDT MC | HDT SC | HDT HAC | HDTC | |
|---|---|---|---|---|---|---|---|---|
| Validity ↑ | 0.618 | 0.884 | 0.396 | 0.128 | 0.892 | 0.068 | 0.144 | 0.918 |
| Uniqueness ↑ | 1.000 | 0.876 | 0.934 | 1.000 | 0.946 | 1.000 | 1.000 | 0.858 |
| Novelty ↑ | 1.000 | 0.756 | 0.914 | 1.000 | 0.859 | 1.000 | 1.000 | 0.802 |
| FCD ↓ | 6.94 | 6.36 | 10.56 | 20.94 | 4.50 | 24.93 | 17.17 | 4.74 |
| SNN ↑ | 0.269 | 0.989 | 0.707 | 0.241 | 0.991 | 0.242 | 0.240 | 0.990 |
| Frag ↑ | 0.944 | 0.948 | 0.926 | 0.800 | 0.965 | 0.754 | 0.885 | 0.969 |
| Scaff ↑ | 0.000 | 0.139 | 0.099 | 0.000 | 0.111 | 0.000 | 0.000 | 0.150 |
| IntDiv ↑ | 0.887 | 0.876 | 0.883 | 0.879 | 0.881 | 0.865 | 0.888 | 0.882 |
| PGD ↓ | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
HDTC achieves the highest validity (91.8%) and fragment similarity (0.969), while HDT MC is a close second (89.2% validity). The flat-walk baseline SENT drops to 61.8% validity — a 30-point gap — showing that flat tokenization struggles with complex molecular graphs. Motif-community (MC) coarsening consistently outperforms both spectral (SC) and agglomerative (HAC) coarsening across all hierarchical tokenizers, confirming the importance of preserving known chemical motifs during coarsening.
| SENT | H-SENT MC | H-SENT SC | H-SENT HAC | HDT MC | HDT SC | HDT HAC | HDTC | |
|---|---|---|---|---|---|---|---|---|
| FG MMD ↓ | 0.004 | 0.003 | 0.006 | 0.018 | 0.002 | 0.031 | 0.015 | 0.003 |
| SMARTS MMD ↓ | 0.005 | 0.006 | 0.014 | 0.039 | 0.002 | 0.066 | 0.028 | 0.004 |
| Ring MMD ↓ | 0.012 | 0.009 | 0.010 | 0.059 | 0.003 | 0.061 | 0.032 | 0.005 |
| BRICS MMD ↓ | 0.046 | 0.051 | 0.052 | 0.087 | 0.037 | 0.097 | 0.065 | 0.037 |
| Motif Rate ↑ | 0.553 | 0.643 | 0.556 | 0.438 | 0.612 | 0.176 | 0.389 | 0.704 |
| Subst. TV ↓ | 0.067 | 0.023 | 0.010 | 0.275 | 0.081 | 0.278 | 0.217 | 0.052 |
| Subst. KL ↓ | 0.018 | 0.002 | 0.000 | 0.362 | 0.016 | 2.509 | 0.179 | 0.008 |
| FG TV ↓ | 0.043 | 0.048 | 0.069 | 0.109 | 0.032 | 0.204 | 0.098 | 0.028 |
| FG KL ↓ | 0.093 | 0.116 | 0.105 | 0.942 | 0.090 | 2.480 | 0.143 | 0.086 |
HDT MC and HDTC dominate across all four MMD metrics. HDTC leads on motif rate (70.4% of valid molecules contain at least one recognized motif) and functional-group fidelity. SC and HAC variants lag significantly, reinforcing that the motif-community and compositional approaches excel at preserving complex substructure distributions in large molecules.
Generation Gallery
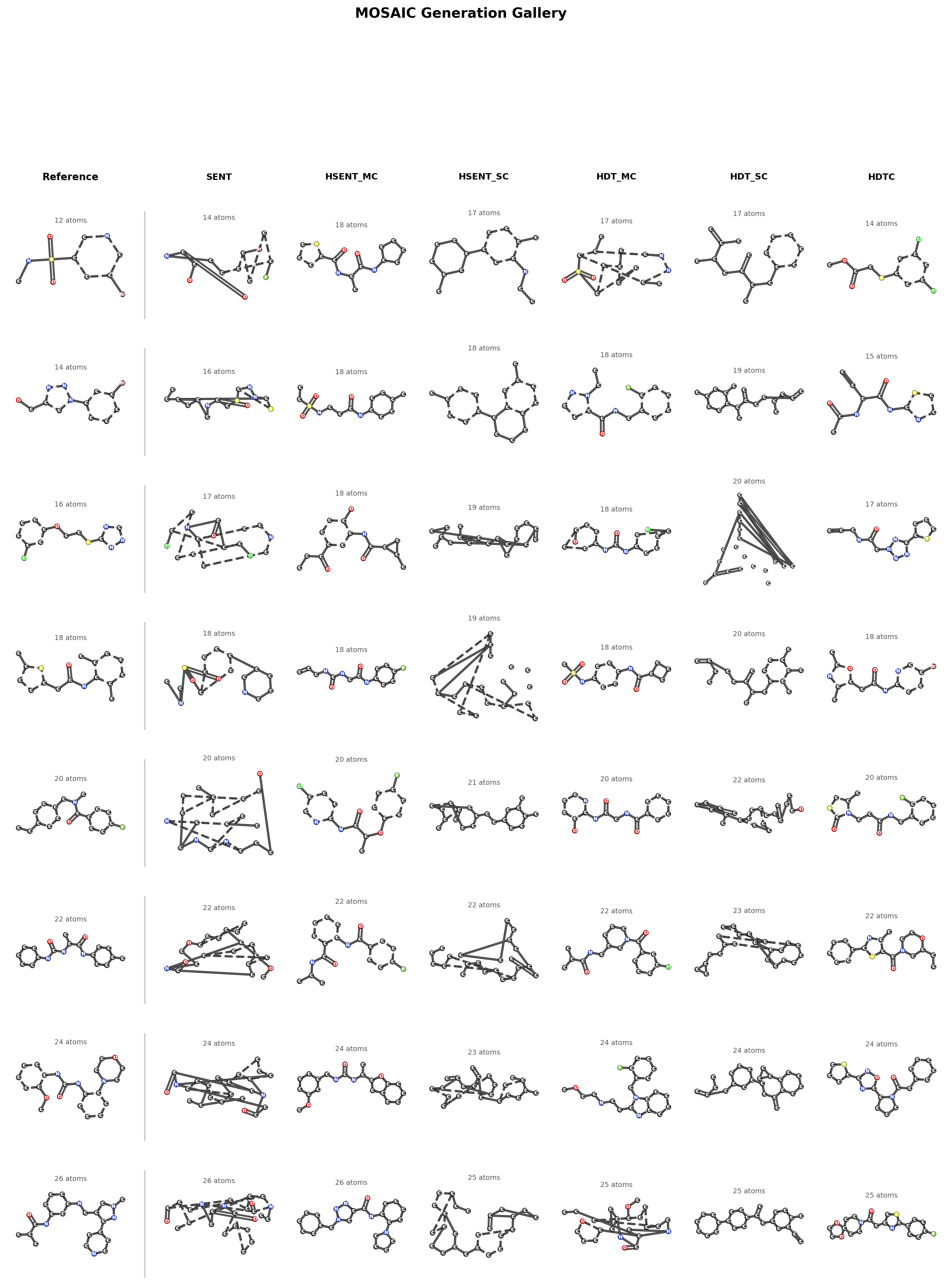
Generation Demo
The following animation shows MOSAIC’s autoregressive generation process, where the model builds a molecule token-by-token, progressively assembling the hierarchical structure from coarse partitions down to individual atoms and bonds.

BibTeX Citation
@article{bian2025mosaic, title = {Beyond Flat Walks: Compositional Abstraction for Autoregressive Molecular Generation}, author = {Bian, Kaiwen and Yang, Andrew H. and Parviz, Ali and Mishne, Gal and Wang, Yusu}, year = {2025}, url = {https://github.com/KevinBian107/MOSAIC},}